Database manipulation with polars¶
You can use any database library to manipulate data in the ARIEL database. In this example we will be using polars.
[ ]:
# Import the libraries
import polars as pl
import sqlite3
import numpy as np
import matplotlib.pyplot as plt
1. How to get data about the experiment¶
As explained above, ARIEL stores every individual that existed during evolution in an SQL database. This includes individuals that died early as well as those that survived until the end. By querying this database, we can recreate population data and extract statistics about the evolutionary process.
1.1 Loading the database into a Polars DataFrame¶
All data about individuals is stored in the table individual. Below is how to load this table into a Polars DataFrame.
[5]:
# Load database table into Polars
conn = sqlite3.connect("__data__/database.db")
data = pl.DataFrame(
conn.execute("SELECT * FROM individual").fetchall(),
schema=[col[1] for col in conn.execute("PRAGMA table_info(individual)")]
)
data.head()
C:\Users\johng\AppData\Local\Temp\ipykernel_21012\3588941998.py:4: DataOrientationWarning: Row orientation inferred during DataFrame construction. Explicitly specify the orientation by passing `orient="row"` to silence this warning.
data = pl.DataFrame(
[5]:
| id | alive | time_of_birth | time_of_death | requires_eval | fitness_ | requires_init | genotype_ | tags_ |
|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | i64 | i64 | f64 | i64 | str | str |
| 1 | 0 | 0 | 7 | 0 | 21.507857 | 0 | "[54.5476194721131, 18.78769761… | "{}" |
| 2 | 0 | 0 | 4 | 0 | 21.764198 | 0 | "[-23.51735411458668, 83.298954… | "{}" |
| 3 | 0 | 0 | 2 | 0 | 21.364822 | 0 | "[-41.8642899864318, -26.625290… | "{}" |
| 4 | 0 | 0 | 4 | 0 | 21.965413 | 0 | "[-2.399348473982663, 32.839368… | "{}" |
| 5 | 0 | 0 | 1 | 0 | 21.85274 | 0 | "[40.97966824037182, -45.669443… | "{}" |
1.2 Reconstructing the population per generation¶
ARIEL does not store a list of individuals per generation. Instead, each individual records:
time_of_birth: the generation it was created
time_of_death: the generation it disappeared
We reconstruct the population for every generation using this information.
[6]:
# Determine generation range
min_gen = int(data["time_of_birth"].min())
max_gen = int(data["time_of_death"].max())
# Collect individuals alive per generation
population_per_gen = {
gen: data.filter(
(pl.col("time_of_birth") <= gen) & (pl.col("time_of_death") > gen) )["id"].to_list()
for gen in range(min_gen, max_gen + 1)
}
# Structure data as a Polars dataframe
pop_df = pl.DataFrame({
"generation": list(population_per_gen.keys()),
"individuals": list(population_per_gen.values()),
"pop size": [len(v) for v in population_per_gen.values()]
})
pop_df.head()
[6]:
| generation | individuals | pop size |
|---|---|---|
| i64 | list[i64] | i64 |
| 0 | [1, 2, … 100] | 100 |
| 1 | [1, 2, … 156] | 100 |
| 2 | [1, 2, … 202] | 100 |
| 3 | [1, 2, … 250] | 100 |
| 4 | [1, 9, … 300] | 100 |
2. Computing fitness statistics per generation¶
Now that we know which individuals were alive in each generation, we can compute:
Mean fitness
Standard deviation
Best (minimum) fitness
[8]:
# Extract fitness values indexed by individual id
fitness_by_id = data.select(["id", "fitness_"])
means, stds, bests = [], [], []
for gen in pop_df["generation"]:
ids = population_per_gen.get(gen, [])
if not ids:
means.append(np.nan)
stds.append(np.nan)
bests.append(np.nan)
continue
fits = (
fitness_by_id
.filter(pl.col("id").is_in(ids))
.select(pl.col("fitness_").cast(pl.Float64))
.to_series()
.to_numpy()
)
if fits.size == 0:
means.append(np.nan)
stds.append(np.nan)
bests.append(np.nan)
else:
means.append(float(np.mean(fits)))
stds.append(float(np.std(fits)))
bests.append(float(np.min(fits)))
# Add the statistics to the dataframe
pop_df = pop_df.with_columns([
pl.Series("fitness_mean", means),
pl.Series("fitness_std", stds),
pl.Series("fitness_best", bests)
])
pop_df.head()
[8]:
| generation | individuals | pop size | fitness_mean | fitness_std | fitness_best |
|---|---|---|---|---|---|
| i64 | list[i64] | i64 | f64 | f64 | f64 |
| 0 | [1, 2, … 100] | 100 | 21.66666 | 0.273939 | 20.806873 |
| 1 | [1, 2, … 156] | 100 | 21.399773 | 0.664911 | 19.846392 |
| 2 | [1, 2, … 202] | 100 | 21.261569 | 0.748077 | 19.952315 |
| 3 | [1, 2, … 250] | 100 | 21.159823 | 0.801033 | 19.862954 |
| 4 | [1, 9, … 300] | 100 | 20.91889 | 0.847252 | 19.862954 |
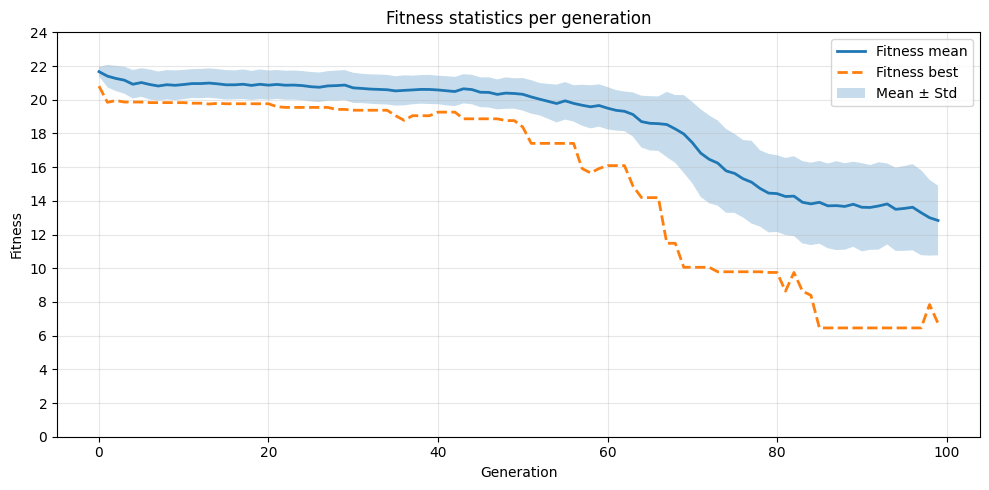
3. Plotting the fitness progression¶
With the computed statistics, we can visualize how fitness changes over generations.
[10]:
df = pop_df.drop_nulls(subset=["fitness_mean"])
x = df["generation"]
mean = df["fitness_mean"]
std = df["fitness_std"]
best = df["fitness_best"]
plt.figure(figsize=(10, 5))
plt.plot(x, mean, label="Fitness mean", linewidth=2)
plt.plot(x, best, "--", label="Fitness best", linewidth=2)
plt.fill_between(x, mean - std, mean + std, alpha=0.25, label="Mean ± Std")
plt.xlabel("Generation")
plt.ylabel("Fitness")
plt.title("Fitness statistics per generation")
plt.legend()
plt.yticks(range(0, int(max(df["fitness_mean"]) + 5), 2))
plt.grid(alpha=0.3)
plt.tight_layout()
plt.show()